Metagenomics
-
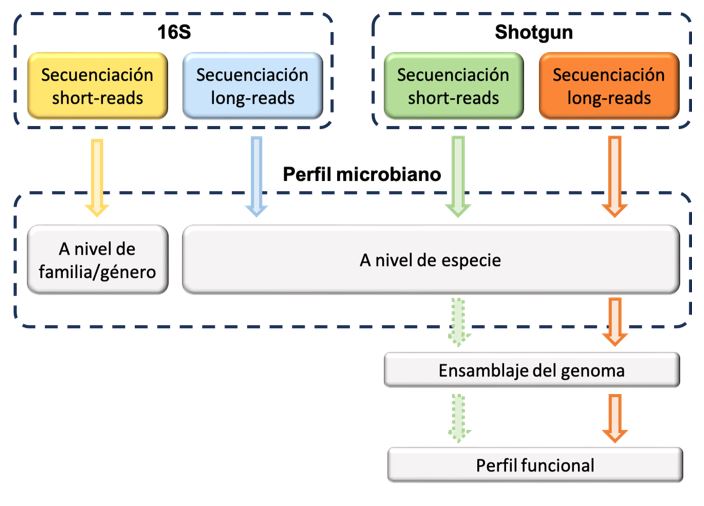
Sequencing of 16S, 18S and ITS regions provides information to differentiate between taxa, establish phylogeny relationships and classify prokaryotic and eukaryotic organisms. In Seqplexing we offer the analysis of the results of specific sequencing of these regions from both short-reads, using amplicons, and long-reads, from the Illumina, PacBIO and ONT platforms.
-
By bioinformatic analysis of the results obtained by random Shotgun sequencing we can detect the bacteria present in a sample, as well as their relative proportion.
In addition, using Shotgun technology from short-reads we can assemble complete genomes from pure cultures. In the case of sequencing with long-reads we can even determine and assemble the complete sequence of the different organisms present in a single sample. -
Sequencing data from metagenomics can be used to obtain the functional profile of the species to be studied and to identify the genes and antibiotic resistance.
-
Results from metatranscriptomics allow us to detect the pathways expressed in the organisms present in a sample.
 ES
ES

